Research
Our group is interested in how protein-protein interactions induce conformational changes at a distance, a phenomenon termed allostery. Allosteric regulation mechanisms that tightly control protein function are widespread in biology, and their disruption is often linked to disease. We use site-specific spectroscopic probes to detect these conformational changes in proteins in solution, and x-ray crystallography to determine their structural nature. Projects in the lab include efforts to discover new regulatory mechanisms of this sort in multi-domain signaling proteins, to understand the evolutionary relationships among allosteric mechanisms within families of proteins, and to design small-molecules that control signaling proteins by modulating their conformation in particular ways.
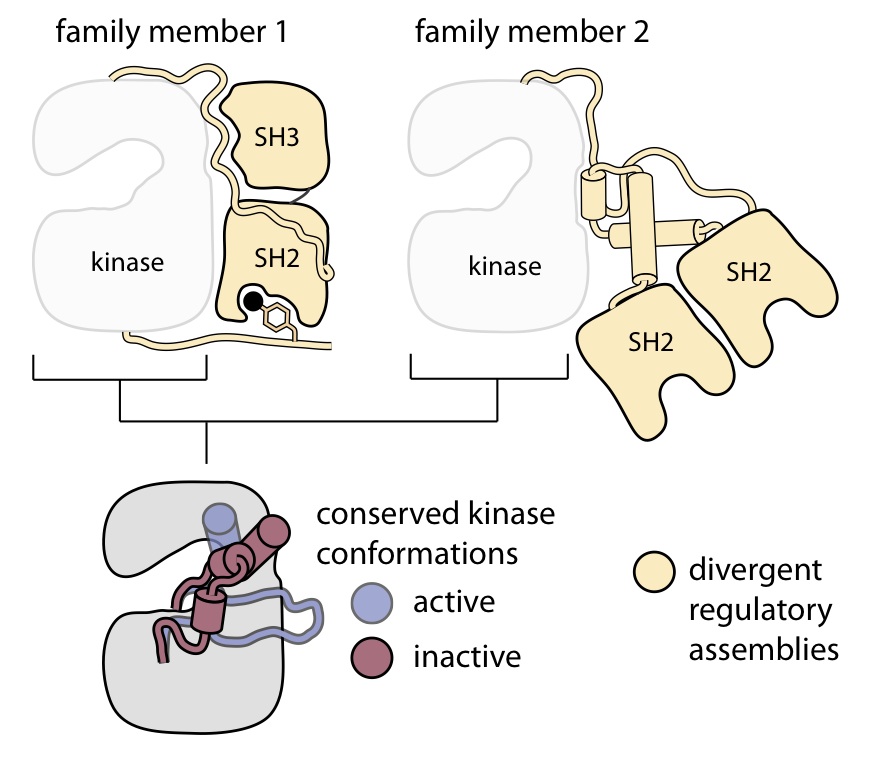
Allostery in protein kinases
The protein kinases are signaling proteins of immense importance to human health, as they are key mediators of cell growth and division. Kinases have intricate regulatory mechanisms that ensure they are only catalytically active when appropriate, but oncogenic kinases typically have mutations that bypass this control, leading to constitutive kinase activity and cancer.
At first glance, kinases appear to have each evolved their own regulatory mechanisms, as the kinase domain is controlled by a diverse set of regulatory domains and subunits in different family members. Nevertheless, the precise conformational changes that the kinase domain undergoes in response to these diverse cues are in fact strikingly conserved. This suggests that all kinase domains have intrinsic allosteric potential that was moulded through evolutionary divergence into a bewildering array of different forms. We are pinpointing where the conservation of allostery gives way to divergence. This question has important practical implications for the selectivity of future allosteric kinase inhibitors, which hold great promise as the cancer drugs of tomorrow.
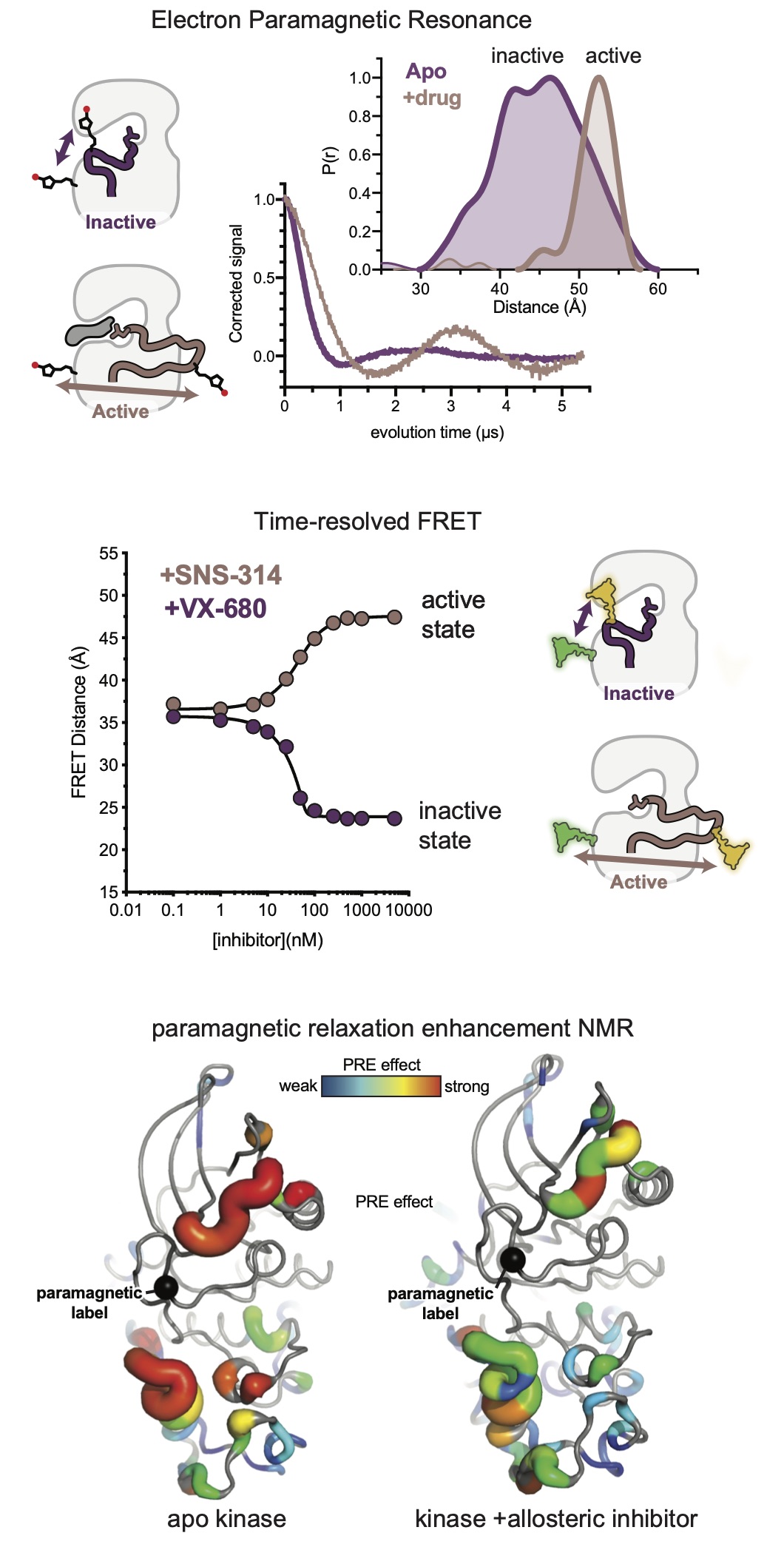
A spectroscopic toolkit for kinase allostery
A critical part of our work is the development of reliable spectroscopic assays for detecting conformational change in a wide range of kinases in solution. We are approaching this problem using a panoply of spectroscopic methods including vibrational spectroscopy, fluorescence and FRET, electron paramagnetic resonance and NMR. Probes are incorporated by chemical modification of the protein or introduced on the chemical scaffold of a kinase inhibitor. These probes provide readouts of the allosteric structural changes triggered by the binding of regulatory proteins or drugs or by posttranslational modifications. Such convenient and direct readouts of kinase conformation have been lacking to date, severely hampering efforts to understand kinase allostery, and their development provides the underlying framework for the research in the lab.
Dissecting allosteric coupling pathways in kinases
X-ray structures hint at how regulatory interactions coax the kinase domain into adopting a particular conformation. What is not clear from such structures is the nature of the allosteric coupling pathways, in other words which elements of the interactions are critical for transmitting the allosteric “signal”. Determining these pathways in multiple kinases will shed light on how the allosteric mechanisms are related in evolutionary terms, and would provide obvious targets for the design of allosteric inhibitors. We plan to delineate these pathways using mutagenesis screens to alter allosteric communication, and spectroscopic methods to readout the effects on kinase conformation.

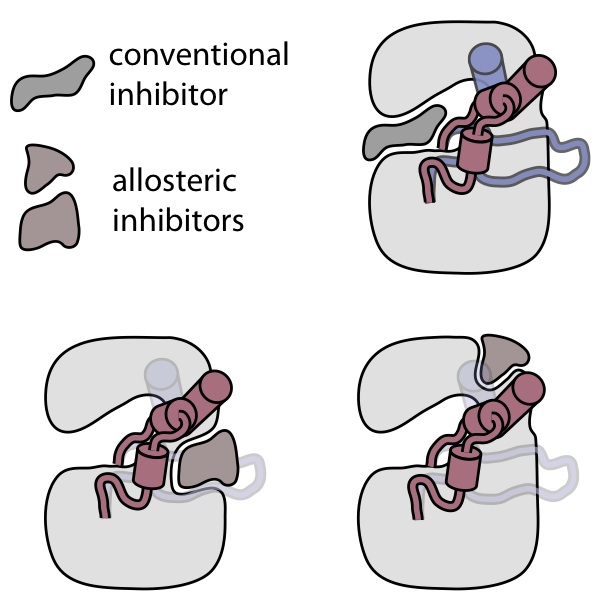
Discovering allosteric kinase inhibitors
Protein kinases are the most important class of drug targets in oncology, and dozens of kinase inhibitors are currently in clinical use. Most of these inhibitors target the highly conserved ATP-binding site, cannot distinguish between different kinase conformations, and suffer from poor selectivity among kinases. Allosteric inhibitors, which block activity by modulating kinase conformation, have proven hard to identify with conventional high-throughput screening approaches. We believe that our spectroscopic conformational assays hold the key for discovering allosteric inhibitors of medically important kinases, which should be much more selective than conventional kinase inhibitors. Our work on allosteric coupling pathways will help us design inhibitors that actually mimic the allosteric control imposed by kinase regulatory domains, essentially restoring oncogenic kinases to their correctly regulated states.
Probing kinase active sites with fluorescent inhibitors
The clinically useful 4-anilinoquinazoline and 4-anilinoquinoline kinase inhibitors are fluorescent molecules whose spectral properties depend on the chemical makeup of the site they are bound to. We are exploiting this convenient property to study the nature of the kinase ATP-binding site and how it differs among related kinases.
© Levinson Lab 2014 | University of Minnesota, Twin Cities | Department of Pharmacology | 312 Church Street SE, Minneapolis, MN 55455
